
Teerapong Buaboocha, Professor, Ph.D.
B.Sc., Chulalongkorn University
Ph.D., University of Illinois
Office: Room 721.01, Science 10 Building
Phone: 662-218-5436
Fax: 662-218-5418
Email: Teerapong.B@chula.ac.th
Research
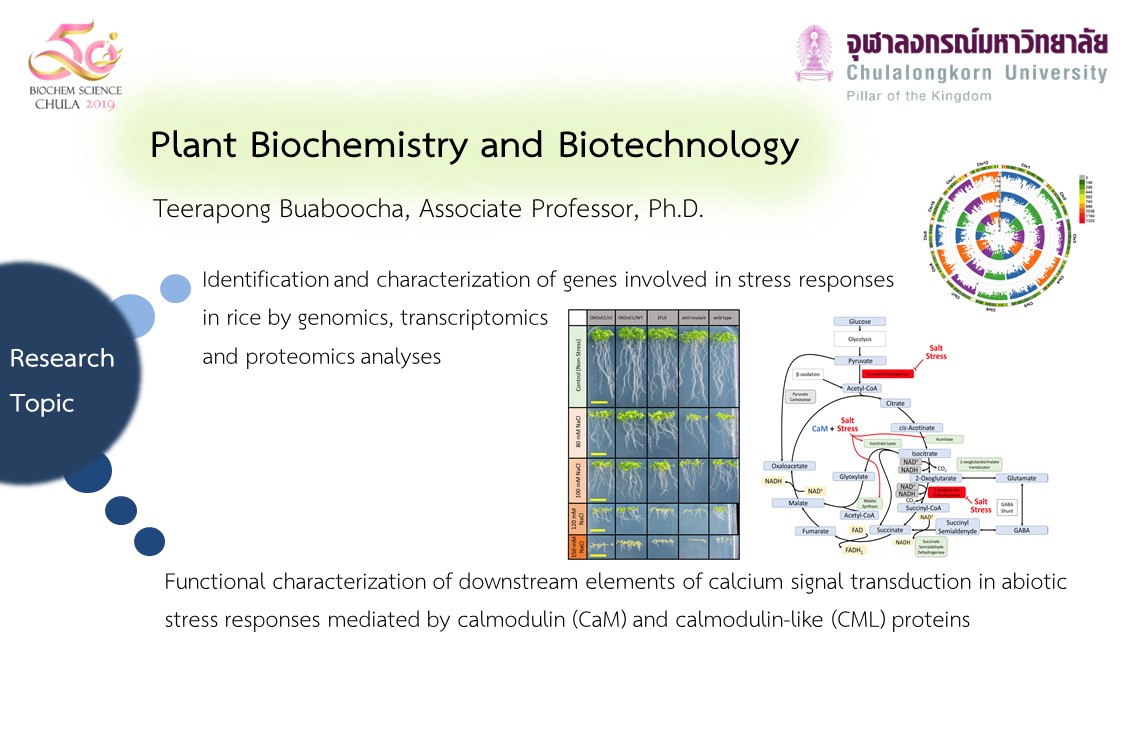 Molecular mechanisms of calcium-mediated stress responses in rice
Molecular mechanisms of calcium-mediated stress responses in rice
I am particularly interested in mechanisms of calcium-mediated responses to stress. Because stress responses are mediated by complex networks of cell signaling, understanding mechanisms of how plants perceive and transmit stress signals and how the functional and regulatory proteins contribute to the tolerance require knowledge of a genome scale. Since the rice genome project has been completed, its genome sequence information can be exploited in this area.
In my lab, we are working on a family of calcium -binding proteins, calmodulin ( CaM ) in rice and trying to determine whether and how they contribute to stress tolerance. We have identified at least 37 CaM and CaM-like (CML) proteins from the rice genome databases. Currently, we focus on the highly conserved CaMs by studying expression of their genes, producing and characterizing the encoded recombinant proteins, identifying their target proteins, producing and examining transgenic rice plants with expression levels of their genes altered.
Our ultimate goal is to understand how stress signals are mediated by calcium-regulated pathways in conjunction with CaM and CML in rice. We will be using proteomic analysis to help identify proteins involved in the calcium/CaM signaling network that mediates stress responses. The knowledge on these proteins not only comprises critical components for understanding stress tolerance, but could also facilitate the effort to genetically improve stress tolerance of crop plants in the future.
Recent Publications
Publications
- Herwibawa B., Lekklar C., Chadchawan S., Buaboocha T. Association of a Specific OsCULLIN3c Haplotype with Salt Stress Responses in Local Thai Rice. International Journal of Molecular Sciences 2024, 25:1040.
- Praphasanobol P., Purnama P.R., Junbuathong S., Chotechuen S., Moung-Ngam P., Kasettranan W., Paliyavuth C., Comai L., Pongpanich M., Buaboocha T., Chadchawan S. Genome-Wide Association Study of Starch Properties in Local Thai Rice. Plants 2023, 12:3290.
- Saputro T.B., Jakada B.H., Chutimanukul P., Comai L., Buaboocha T., Chadchawan S. OsBTBZ1 Confers Salt Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences 2023, 24:14483.
- Kanchitanurak P., Chadchawan S., Buaboocha T. Effects of calmodulin overexpression on gamma-aminobutyric acid (GABA) levels and glutamate decarboxylase activity in rice seedlings. ScienceAsia 2023, 49:646-652.
- Thanabut, S., Sornplerng, P., Buaboocha, T. Ectopic expression of rice malate synthase in Arabidopsis revealed its roles in salt stress responses. Journal of Plant Physiology 2023, 280:153863.
- Sangchai, P., Buaboocha, T., Sirikantaramas, S., Wutipraditkul, N. Changes in physiological responses of OsCaM1-1 overexpression in the transgenic rice under dehydration stress. Bioscience, Biotechnology, and Biochemistry 2022, 86:1211-1219.
- Pinit, S., Ruengchaijatuporn, N., Sriswasdi, S., Buaboocha, T., Chadchawan, S., Chaiwanon, J. Hyperspectral and genome-wide association analyses of leaf phosphorus status in local Thai indica rice. PLoS ONE 2022, 17:e0267304.
- Habila, S., Khunpolwattana, N., Chantarachot, T., Buaboocha, T., Comai, L., Chadchawan, S., Pongpanich, M. Salt stress responses and SNP-based phylogenetic analysis of Thai rice cultivars. Plant Genome 2022, e20189.
- Kojonna, T., Suttiyut, T., Khunpolwattana, N., Pongpanich, M., Suriya‐arunroj, D., Comai, L., Buaboocha, T., Chadchawan, S. Identification of a negative regulator for salt tolerance at seedling stage via a genome‐wide association study of Thai rice populations. International Journal of Molecular Sciences 2022, 23:1842.
- Sonsungsan, P., Chantanakool, P., Suratanee, A., Buaboocha, T., Comai, L., Chadchawan, S., Plaimas, K. Identification of Key Genes in ‘Luang Pratahn’, Thai Salt-Tolerant Rice, Based on Time-Course Data and Weighted Co-expression Networks. Frontiers in Plant Science 2021, 12:744654.
- Iqbal, Z., Iqbal, M.S., Sangpong, L., Khaksar, G., Sirikantaramas, S., Buaboocha, T. Comprehensive genome-wide analysis of calmodulin-binding transcription activator (CAMTA) in Durio zibethinus and identification of fruit ripening-associated DzCAMTAs. BMC Genomics (2021), 22:743.
- Chutimanukul P, Saputro TB, Mahaprom P, Plaimas K, Comai L, Buaboocha T, Siangliw M, Toojinda T, Chadchawan S. Combining genome and gene co-expression network analyses for the identification of genes potentially regulating salt tolerance in rice. Frontiers in Plant Science 2021, 12:704549.
- Suratanee A, Buaboocha T, Plaimas K. Prediction of human-Plasmodium vivax protein associations from heterogeneous network structures based on machine-learning approach. Bioinformatics and Biology Insights 2021, 15.
- Iqbal Z, Shariq Iqbal M, Singh SP, Buaboocha T. Ca2+/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Frontiers in Plant Science 2020, 11:598327.
- Punchkhon C, Plaimas K, Buaboocha T, Siangliw, JL, Toojinda T, Comai L, Diego ND, Spichal L, Chadchawan S. Drought-tolerance gene identification using genome comparison and co-expression network analysis of chromosome substitution lines in rice. Genes 2020, 11:1197.
- Khrueasan N, Siangliw M, Toojinda T, Imyim, A, Buaboocha T, Chadchawan S. Physiological mechanisms of the seedling stage salt tolerance of near isogenic rice lines with the ‘KDML105’ genetic background. International Journal of Agriculture and Biology 2020, 23:927-934.
- Yuenyong W, Sirikantaramas S, Qu L-J, Buaboocha T. Isocitrate lyase plays important roles in plant salt tolerance. BMC Plant Biology 2019, 19:472.
- Khrueasan N, Chutimanukul P, Plaimas K, Buaboocha T, Siangliw M, Toojinda T, Comai L, Chadchawan S. Comparison between the transcriptomes of ‘KDML105′ rice and a salt-tolerant chromosome segment substitution line. Genes 2019, 10:742.
- Lekklar C, Suriya-arunroj D, Pongpanich M, Comai L, Kositsup B., Chadchawan, S, Buaboocha T. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes 2019, 10:562.
- Lekklar C, Pongpanich M, Suriya-arunroj D, Chinpongpanich A, Tsai H, Comai L, Chadchawan S, Buaboocha T. Genome-wide association study for salinity tolerance at the flowering stage in a panel of rice accessions from Thailand. BMC Genomics 2019 20:76.
- Kaewneramit T, Buaboocha T, Sangchai P, Wutipraditkul N. OsCaM1-1 overexpression in the transgenic rice mitigated salt-induced oxidative damage. Biologia Plantarum 2019, 63:335-342.
- Yuenyong W, Chinpongpanich A, Phean-o-pas S, Comai L, Chadchawan S, Buaboocha T. Downstream components of calmodulin signaling pathway in rice salt stress response revealed by transcriptome profiling and target identification. BMC Plant Biology 2018, 18:335.
- Suratnee A, Chokrathok C, Chutimanukul P, Khrueasan N, Buaboocha T, Chadchawan S, Plaimas K. Two-state co-expression network analysis to identify genes related to salt tolerance in Thai rice. Genes 2018, 9:594.
- Boonchai C, Udomchalothorn T, Sripinyowanich S, Comai L, Bauboocha T, Chadchawan S. Rice overexpressing OsNUC1-S reveals differential gene expression leading to yield loss reduction after salt stress at the booting stage. International Journal of Molecular Sciences 2018, 19:3936.
- Chutimanukul P, Kositsup B, Plaimas K, Buaboocha T, Siangliw M, Toojinda T, Comai L, Chadchawan S. Data in support of photosynthetic responses in a chromosome segment substitution line of ‘Khao Dawk Mali 105’ rice at seedling stage. Data in Brief 2018, 21: 307-312.
- Chutimanukul P, Kositsup B, Plaimas K, Buaboocha T, Siangliw M, Toojinda T, Comai L, Chadchawan S. Photosynthetic responses and identification of salt tolerance genes in a chromosome segment substitution line of ‘Khao Dawk Mali 105’ rice. Environ Exp Bot 2018, 155: 497-508.
- Chaicherdsakul T, Yuenyong W, Roytrakul S, Chadchawan S, Wutiprsditkul N, Limpaseni T, Buaboocha T. Proteomic analysis of transgenic rice overexpressing a calmodulin calcium sensor reveals its effects on redox signaling and homeostasis. J Plant Biochem Biotechnol 2017, 26: 235-245.
- Udomchalothorn T, Plaimas K, Sripinyowanich S, Boonchai C, Kojonna T, Chutimanukul P, Comai L, Buaboocha T, Chadchawan S. OsNucleolin1-L expression in arabidopsis enhances photosynthesis via transcriptome modification under salt stress conditions. Plant Cell Physiol 2017, 58: 717-734.
- Chinpongpanich A, Phean-o-pas S, ThongchuangM, Qu Li-Jia, Buaboocha T. C-Terminal Extension of Calmodulin-Like 3 Protein from Oryza sativa L.: Interaction with a High Mobility Group Target Protein. Acta Bioch Bioph Sin 2015, 47: 880-889.
- Wutipraditkul N, Wongwean P, Buaboocha T. Alleviation of salt-induced oxidative stress in rice seedlings by proline and/or glycinebetaine. Biologia Plantarum 2015, 59: 547-553.
- Udomchalothorn T, Plaimas K, Comai L, Buaboocha T, Chadchawan S. Molecular karyotyping and exome Analysis of salt-Tolerant rice mutant from somaclonal variation. Plant Genome 2014, 7:3.
- Seang-Ngam S, Limruengroj K, Pichyangkura R, Chadchawan S, Buaboocha T. Chitosan potentially induces drought resistance in rice Oryza sativa L. via calmodulin. J Chitin Chitosan Sci, 2014, 117-122.
- Sripinyowanich S, Chamnanmanoontham N, Udomchalothorn T, Maneeprasopsuk S, Santawee P, Buaboocha T, Qu L-J, Gu H, Chadchawan S. Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Science, 2013, 213: 67-78.
- Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu H, Buaboocha T, Chadchawan S. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot 2013, 86: 94-105.
- Chinpongpanich A, Limruengroj K, Phean-o-pas S, Limpaseni T, Buaboocha T. Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Research Notes 2012, 5:625.
- Saeng-ngam S, Takpirom W, Buaboocha T, Chadchawan S. The role of the OsCam1-1 salt stress in ABA accumulation and salt tolerance in rice. J Plant Biol 2012, 55: 198-208.
- Chinpongpanich A, Wutipraditkul N, Thairat S, Buaboocha T. Biophysical characterization of calmodulin and calmodulin-like proteins from rice, Oryza sativa L. Acta Bioch Bioph Sin 2011, 43: 867-876.
- Wutipraditkul N, Boonkomrat S, Buaboocha T. Expression and characterization of catalases from rice, Oryza sativa. Biosci Biotech Bioch 2011, 75(10): 1900-1906.
- Phean-o-pas S, Limpaseni T, Buaboocha T. Structure and expression analysis of the OsCam1-1 calmodulin gene from Oryza sativa L. BMB Rep 2008, 41(11): 771-777.
- Boonburapong B, Buaboocha T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biology 2007, 7:4
- Laloknam S, Tanaka K, Buaboocha T, Waditee R, Incharoensakdi A, Hibino T, Tanaka Y, Takabe T. A Halotolerant cyanobacterium Aphanothece halophytica contains betaine transporter that is active at alkaline pH under high salinity conditions. Appl Environ Microb 2006, 72(9): 6018-6026.
- Waditee R, Buaboocha T, Kato M, Hibino T, Suzuki S, Nakamura T, Takabe T. Carboxyl terminal hydrophilic tail of a NhaP type Na+/H+ antiporter from cyanobacteria is involved in the apparent affinity for Na+ and pH sensitivity. Arch Biochem Biophys 2006, 450(1): 113-121.
- Phean-o-pas S, Punteeranurak P, Buaboocha T. Calcium signaling-mediated and differential induction of calmodulin gene expression by stress in Oryza sativa L. J Biochem Mol Biol 2005, 38(4): 432-439.
- Buaboocha T, Zielinski RE. Calmodulin In: Annual Plant Reviews, Vol.7, Protein-Protein Interactions (MT. McManus, W. Liang and A. Allen, eds.) Sheffield Academic Press, Sheffield, UK. 2002, 285-313.
- Buaboocha T, Liao B, Zielinski RE. Isolation of cDNA and genomic DNA clones encoding a calmodulin-binding protein related to a family of ATPases involved in cell division and vesicle fusion. Planta 2001, 212: 774-781.
Books
ธีรพงษ์ บัวบูชา. จีโนมิกส์ของพืช. พิมพ์ครั้งที่ 1. กรุงเทพฯ : โรงพิมพ์แห่งจุฬาลงกรณ์มหาวิทยาลัย, 2552. 254 หน้า.

