
Kittikhun Wangkanont, Ph.D.
B.S., University of Wisconsin-Madison
A.M., Harvard University
Ph.D., University of Wisconsin-Madison
Research
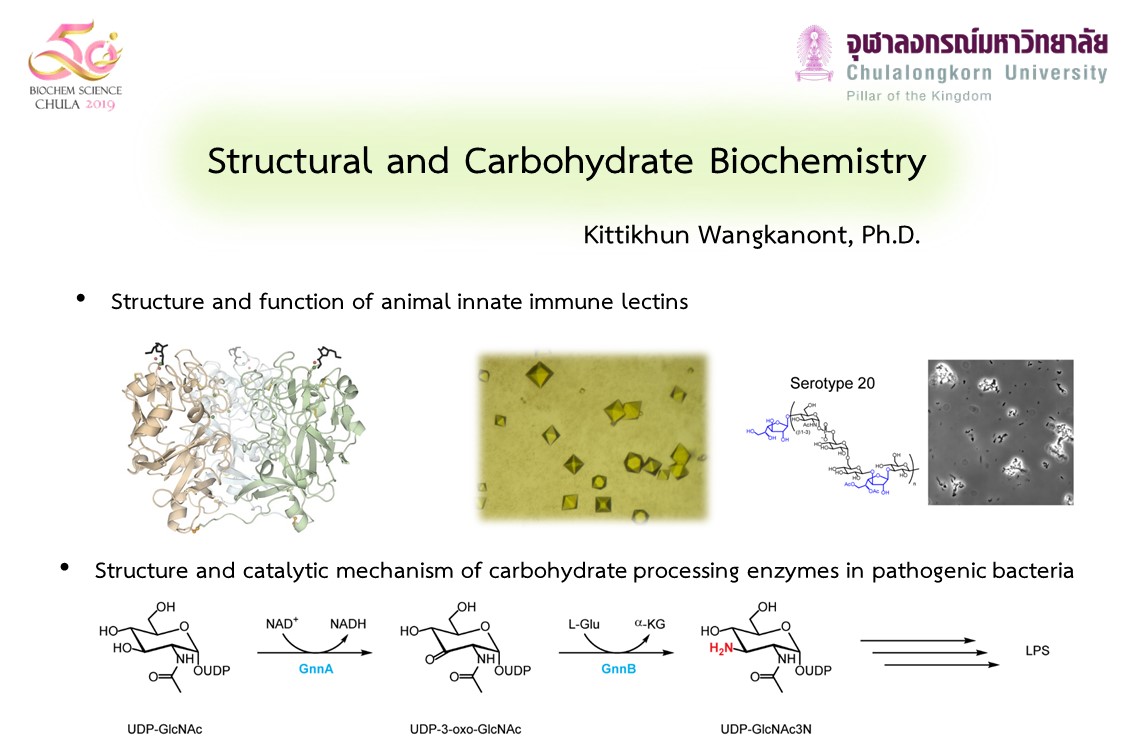
In addition to their usage as energy sources, carbohydrates serve as molecular markers for cellular recognition processes, especially in distinguishing host cells and invading microbes. We are interested in the molecular mechanism of bacteria recognition by carbohydrate binding proteins of the innate immune system as well as the carbohydrate biosynthetic enzymes and virulence factors in pathogenic bacteria.
Carbohydrate binding proteins or lectins are ubiquitous in nature. We focus on characterization of novel intelectins, ficolins, and C-type lectins in the animal innate immune system. Several of these lectins recognize microbes and initiate the immune response. We currently aim to determine carbohydrate and microbe specificity of immune lectins in shrimp and fish species. Immune lectins in reptiles and silkworms are also on the horizon. We envision that these lectins could be used in microbe detection, microbe identification, and development of novel microbe-targeting therapeutics.
In addition to lectins, we are investigating carbohydrate biosynthetic enzymes crucial for lipopolysaccharide assembly in Leptospira interrogans and Burkholderia pseudomallei. Moreover, we are also characterizing metabolic enzymes and virulence factors that are drug targets in these bacteria. We aim to elucidate the catalytic mechanism of these enzymes as well as obtaining crystal structures in complex with inhibitors. The ultimate goal is to obtain novel drug development candidates.
The main focus of our research group is biophysics, structural biology, and chemical biology of host-microbe interactions. The techniques employ in our laboratory include X-ray crystallography, small-angle X-ray scattering, ligand binding analysis, enzyme kinetics, and structure-based inhibitor design. We also routinely produce recombinant proteins in bacteria, yeasts, insect cells, and mammalian cells.
Prospective students and researchers who are interested in the experimental aspects of carbohydrate recognition and enzyme inhibitors are encouraged to contact Dr. Kittikhun directly.
Recent Publications
Publications
- Wangkanont K.; Wesener DA.; Vidani JA.; Kiessling LL.; Forest KT.. Structures of Xenopus Embryonic Epidermal Lectin Reveal a Conserved Mechanism of Microbial Glycan Recognition. J Biol Chem. 2016, 291 (11), 5596-5610.
- Kincaid VA.; London N.; Wangkanont K.; Wesener DA.; Marcus SA.; Héroux A.; Nedyalkova L.; Talaat AM.; Forest KT.; Shoichet BK.; Kiessling LL.. Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents. ACS Chem Biol. 2015, 10 (10), 2209-2218.
- Wesener DA.; Wangkanont K.; McBride R.; Song X.; Kraft MB.; Hodges HL.; Zarling LC.; Splain RA.; Smith DF.; Cummings RD.; Paulson JC.; Forest KT.; Kiessling LL.. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol. 2015, 22 (8), 603-610.
- Wangkanont K.; Forest KT.; Kiessling LL.. The non-detergent sulfobetaine-201 acts as a pharmacological chaperone to promote folding and crystallization of the type II TGF-β receptor extracellular domain. Protein Expr Purif 2015. 115, 19-25.
- Burgett AW.; Poulsen TB.; Wangkanont K.; Anderson DR.; Kikuchi C.; Shimada K.; Okubo S.; Fortner KC.; Mimaki Y.; Kuroda M.; Murphy JP.; Schwalb DJ.; Petrella EC.; Cornella-Taracido I.; Schirle M.; Tallarico JA.; Shair MD.. Natural products reveal cancer cell dependence on oxysterol-binding proteins.
Nat Chem Biol 2011. , 7 (9), 639-647. - Garber KC.; Wangkanont K.; Carlson EE.; Kiessling LL.. A general glycomimetic strategy yields non-carbohydrate inhibitors of DC-SIGN. Chem Commun (Camb) 2010. , 46 (36), 6747-6749.
- Allen MJ.; Wangkanont K.; Raines RT.; Kiessling LL.. ROMP from ROMP: A New Approach to Graft Copolymer Synthesis. Macromolecules 2009. , 42 (12), 4023-4027.

